Data Science Example - Papers about Data Science
About
This is an example of a notebook to demonstrate concepts of Data Science. In this example we will do some very basic plotting of a publications dataset.
We will use some basic data extracted from the DBLP. The dataset contains three fields: a character (either 'A' for article in a journal or 'P' for publication in a conference), the title of the publication and the year it was published. There are more 3.9 million entries in the dataset. You can download the dataset here (330MB).
Let's import the libraries we will need:
library(data.table) library(ggplot2)
Reading the data
We can read a CSV file into R with the following code. It will parse the CSV file, using the header (first line) to name the fields and keep strings as strings.
data <- read.csv("Data/OldPubs/articles.csv", header = TRUE,stringsAsFactors=FALSE)
Let's see the structure of the dataframe:
str(data)
## 'data.frame': 3969412 obs. of 3 variables: ## $ Type : chr "A" "A" "A" "A" ... ## $ Title: chr "Mobeacon: An iBeacon-Assisted Smartphone-Based Real Time Activity Recognition Framework." "An interactive visualization framework for performance analysis." "Vertical Navigation of Layout Adapted Web Documents." "AO4BPEL: An Aspect-oriented Extension to BPEL." ... ## $ Year : int 2015 2015 2007 2007 2007 2007 2007 2007 2007 2007 ...
Field Type could be a factor, but we don't plan to use it in this application, let's leave it as it is.
Counting papers with specific words in their titles
We want to see whether specific words and combinations appear in the papers' titles. Let's use a naive approach: grep. We'll just create data frames where the titles are the ones that matches the string passed as first parameter to grep.
filteredDS <- data[grep("Data Science", data$Title,ignore.case=TRUE), ] nrow(filteredDS)
## [1] 377
Let's try with other keywords!
filteredDM <- data[grep("Data Mining", data$Title,ignore.case=TRUE), ] nrow(filteredDM)
## [1] 8186
filteredBD <- data[grep("Big Data", data$Title,ignore.case=TRUE), ] nrow(filteredBD)
## [1] 6644
filteredAI <- data[grep("Artificial Intelligence", data$Title,ignore.case=TRUE), ] nrow(filteredAI)
## [1] 2851
filteredNN <- data[grep("Neural Network", data$Title,ignore.case=TRUE), ] nrow(filteredNN)
## [1] 48475
filteredML <- data[grep("Machine Learning", data$Title,ignore.case=TRUE), ] nrow(filteredML)
## [1] 8337
grep works, but we need to know how many papers with a specific word on its title were published by year. table can help us there.
table(filteredDS$Year)
## ## 2001 2006 2007 2008 2009 2010 2011 2012 2013 2014 2015 2016 2017 2018 ## 1 1 1 1 9 1 4 6 10 43 69 88 122 21
Looks OK, but we would like to have the count results as dataframes with the proper column names. Let's create tables for each subset of the data, convert them to dataframes, relabel these dataframes and ensure that the year is treated as a value (instead of a factor).
DSFrequency <- as.data.frame(table(filteredDS$Year)) names(DSFrequency) <- c("Year","Data Science") DSFrequency$Year <- as.numeric(levels(DSFrequency$Year))[DSFrequency$Year] DSFrequency
## Year Data Science ## 1 2001 1 ## 2 2006 1 ## 3 2007 1 ## 4 2008 1 ## 5 2009 9 ## 6 2010 1 ## 7 2011 4 ## 8 2012 6 ## 9 2013 10 ## 10 2014 43 ## 11 2015 69 ## 12 2016 88 ## 13 2017 122 ## 14 2018 21
Much better! Let's do the same for the other dataset subsets.
DMFrequency <- as.data.frame(table(filteredDM$Year)) names(DMFrequency) <- c("Year","Data Mining") DMFrequency$Year <- as.numeric(levels(DMFrequency$Year))[DMFrequency$Year] BDFrequency <- as.data.frame(table(filteredBD$Year)) names(BDFrequency) <- c("Year","Big Data") BDFrequency$Year <- as.numeric(levels(BDFrequency$Year))[BDFrequency$Year] AIFrequency <- as.data.frame(table(filteredAI$Year)) names(AIFrequency) <- c("Year","Artificial Intelligence") AIFrequency$Year <- as.numeric(levels(AIFrequency$Year))[AIFrequency$Year] NNFrequency <- as.data.frame(table(filteredNN$Year)) names(NNFrequency) <- c("Year","Neural Networks") NNFrequency$Year <- as.numeric(levels(NNFrequency$Year))[NNFrequency$Year] MLFrequency <- as.data.frame(table(filteredML$Year)) names(MLFrequency) <- c("Year","Machine Learning") MLFrequency$Year <- as.numeric(levels(MLFrequency$Year))[MLFrequency$Year]
We need to merge all these dataframes together. See Simultaneously merge multiple data.frames in a list for some ways to do that.
all <- Reduce(function(dtf1, dtf2) merge(dtf1,dtf2,by="Year",all=TRUE), list(DSFrequency,DMFrequency,BDFrequency,AIFrequency,NNFrequency,MLFrequency)) head(all)
## Year Data Science Data Mining Big Data Artificial Intelligence ## 1 1959 NA NA NA NA ## 2 1962 NA NA NA 3 ## 3 1963 NA NA NA 1 ## 4 1964 NA NA NA 1 ## 5 1966 NA NA NA 1 ## 6 1968 NA NA NA 1 ## Neural Networks Machine Learning ## 1 NA 2 ## 2 NA 1 ## 3 NA 1 ## 4 NA 1 ## 5 NA 1 ## 6 1 NA
This is a bit messy -- there are a lot of NAs caused by the merging of the dataframes. Let's fix this (here's how: How do I replace NA values with zeros in an R dataframe?):
all[is.na(all)] <- 0 str(all)
## 'data.frame': 56 obs. of 7 variables: ## $ Year : num 1959 1962 1963 1964 1966 ... ## $ Data Science : num 0 0 0 0 0 0 0 0 0 0 ... ## $ Data Mining : num 0 0 0 0 0 0 0 0 0 0 ... ## $ Big Data : num 0 0 0 0 0 0 0 0 0 0 ... ## $ Artificial Intelligence: num 0 3 1 1 1 1 4 2 8 3 ... ## $ Neural Networks : num 0 0 0 0 0 1 0 0 0 0 ... ## $ Machine Learning : num 2 1 1 1 1 0 1 0 0 0 ...
There are also papers published before 1980, let's remove those. Let's also remove data for 2018 since it is incomplete.
all <- all[all$Year >= 1980, ] all <- all[all$Year < 2018, ]
We're ready to plot the time series! Since we want one of the lines to be thicker than the others, we need an auxiliary field (see When creating a multiple line plot in ggplot2, how do you make one line thicker than the others?). See also ggplot2: axis manipulation and themes.
melted <- melt(all,id="Year") colnames(melted) <- c("Year", "Keyword","Count") melted$thickness <- 1 melted$thickness[melted$Keyword=="Data Science"] <- 3 head(melted)
## Year Keyword Count thickness ## 1 1980 Data Science 0 3 ## 2 1981 Data Science 0 3 ## 3 1982 Data Science 0 3 ## 4 1983 Data Science 0 3 ## 5 1984 Data Science 0 3 ## 6 1985 Data Science 0 3
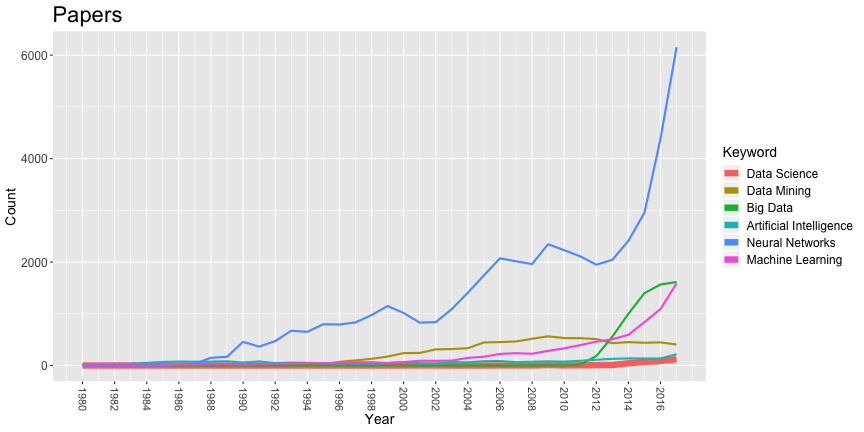
ggplot(melted,aes(x=Year,y=Count,colour=Keyword,group=Keyword,size=thickness)) + geom_line()+ scale_size(range = c(1,3), guide="none")+ scale_x_continuous("Year", breaks=seq(1980,2017,2))+ guides(colour = guide_legend(override.aes = list(size=3)))+ ggtitle("Papers")+ theme(axis.title=element_text(size=14), axis.text.x=element_text(size=11,angle=-90,vjust=0.5,hjust=1), axis.text.y=element_text(size=12), legend.title=element_text(size=14), legend.text=element_text(size=12), plot.title=element_text(size=22))
Exercises
Here are some suggested exercises:
- Our simple grep use is not a very good choice: we didn't consider case variations and may not get titles similar to what could be interesting (e.g. "Data Scientist"). Try a better approach to filter books by title.
- Try different keywords in different domains to see changes in publications' themes.
Warning: Code and results presented on this document are for reference use only. Code was written to be clear, not efficient. There are several ways to achieve the results, not all were considered.
See the R source code for this notebook.